Evaluating coverage bias in next-generation sequencing of Escherichia coli
4.9 (535) In stock
Whole-genome sequencing is essential to many facets of infectious disease research. However, technical limitations such as bias in coverage and tagmentation, and difficulties characterising genomic regions with extreme GC content have created significant obstacles in its use. Illumina has claimed that the recently released DNA Prep library preparation kit, formerly known as Nextera Flex, overcomes some of these limitations. This study aimed to assess bias in coverage, tagmentation, GC content, average fragment size distribution, and de novo assembly quality using both the Nextera XT and DNA Prep kits from Illumina. When performing whole-genome sequencing on Escherichia coli and where coverage bias is the main concern, the DNA Prep kit may provide higher quality results; though de novo assembly quality, tagmentation bias and GC content related bias are unlikely to improve. Based on these results, laboratories with existing workflows based on Nextera XT would see minor benefits in transitioning to the DNA Prep kit if they were primarily studying organisms with neutral GC content.
Molecular identification of plants: from sequence to species

Number of blind spots uniquely present or absent in each sequencing

In silico evaluation and selection of the best 16S rRNA gene primers for use in next-generation sequencing to detect oral bacteria and archaea, Microbiome
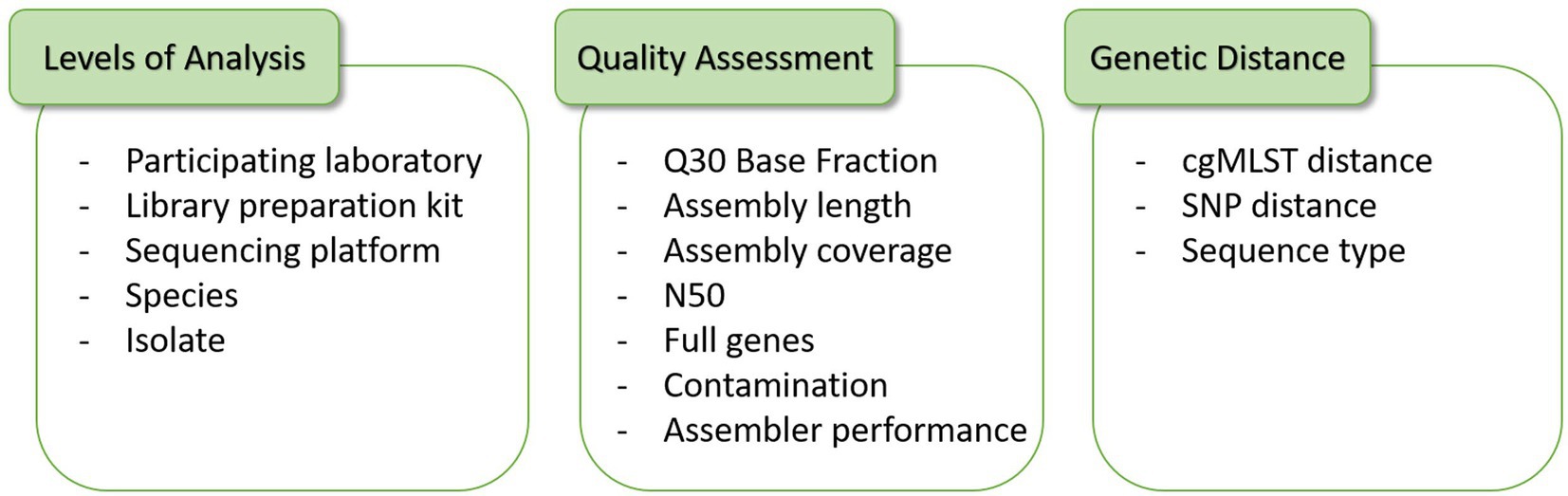
Frontiers Impact of wet-lab protocols on quality of whole-genome short-read sequences from foodborne microbial pathogens
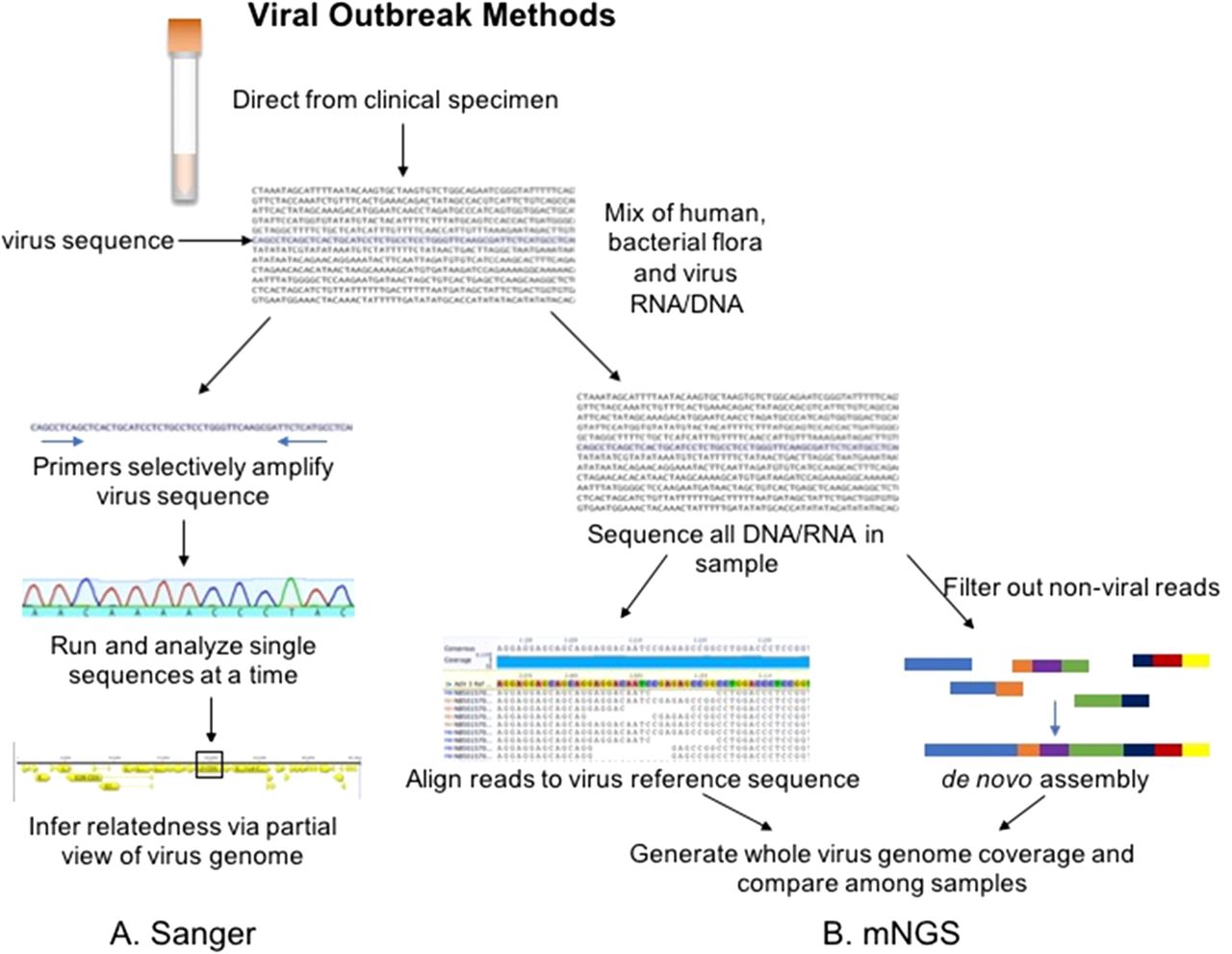
Next-generation Sequencing for Outbreak Investigation in the Clinical Microbiology Laboratory

Nanopore sequencing and its application to the study of microbial communities - ScienceDirect

Pathogens, Free Full-Text

PDF] Summarizing and correcting the GC content bias in high-throughput sequencing
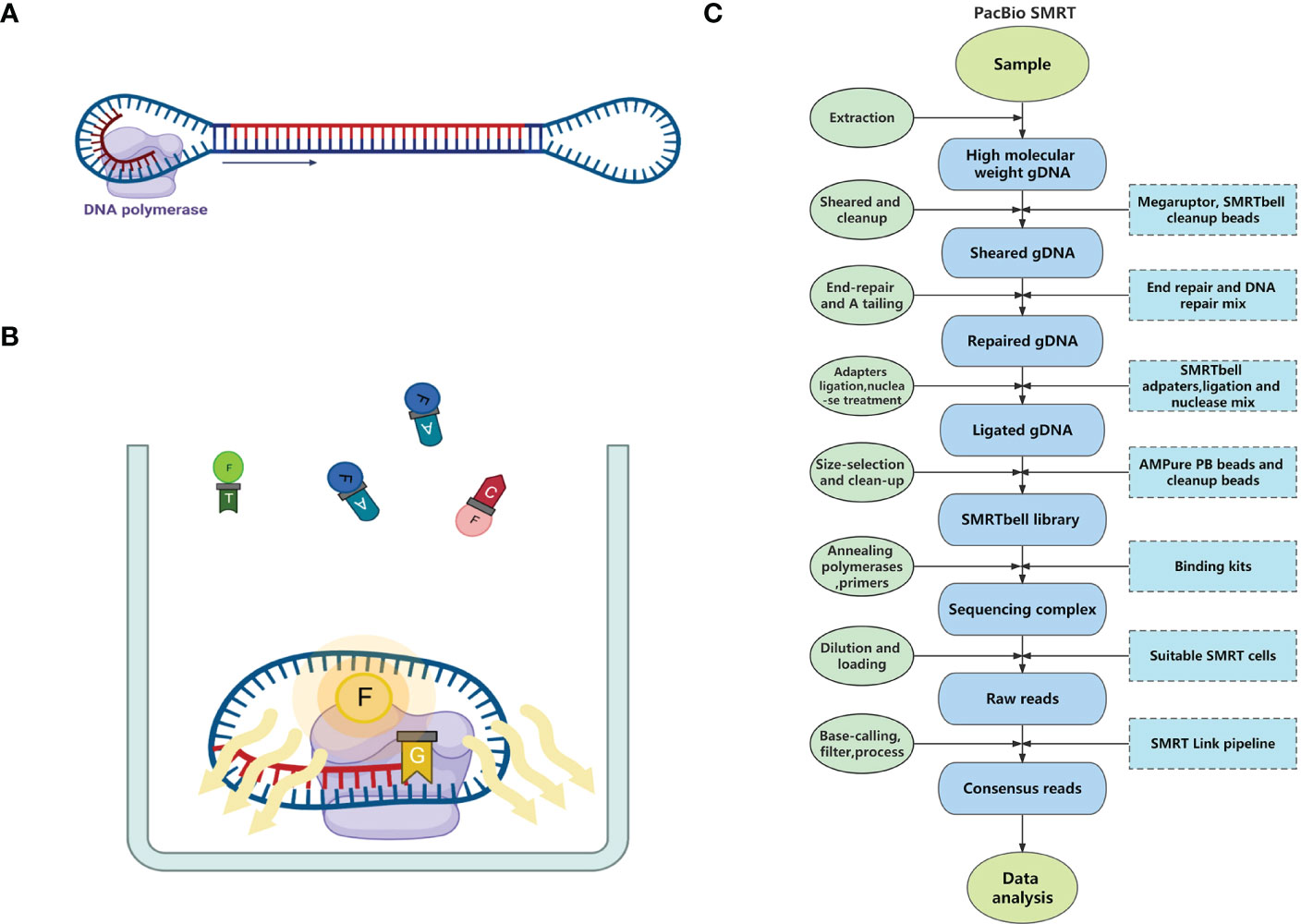
Frontiers Application of third-generation sequencing to herbal genomics

A framework for assessing 16S rRNA marker-gene survey data analysis methods using mixtures., Microbiome

Single cell sequencing - MyBioSource Learning Center

PDF] Illuminating Choices for Library Prep: A Comparison of Library Preparation Methods for Whole Genome Sequencing of Cryptococcus neoformans Using Illumina HiSeq
Low-E glass affect 5G/4G wireless coverage
A protocol for applying low-coverage whole-genome sequencing data
 6 Pcs/Lot Disney Frozen Snow White Princess Cartoon Young Girl
6 Pcs/Lot Disney Frozen Snow White Princess Cartoon Young Girl How To Rock The Double Dutch Braid - Number 4 High Performance
How To Rock The Double Dutch Braid - Number 4 High Performance Compra online de Sutiã feminino macio sem costura com lingerie de
Compra online de Sutiã feminino macio sem costura com lingerie de 44DDD , White : Seamless Full Figure Minimizer Bra
44DDD , White : Seamless Full Figure Minimizer Bra New York Post on X: Model spends $25K on surgery to increase
New York Post on X: Model spends $25K on surgery to increase Calça Academia Fitness Legging Levanta Bumbum Sem Costura (POPPY) - Halifax Fit
Calça Academia Fitness Legging Levanta Bumbum Sem Costura (POPPY) - Halifax Fit